Performance of NAMD will be different for different systems you are simulating, depending especially on the number of atoms in the simulation. Collecting and providing this kind of data is also very useful if you are applying for a RAC award. Even software not listed as available on an HPC cluster is generally available on the login nodes of the cluster assuming it is available for the appropriate OS version; e. These older versions are NOT supported on Deepthought2. These provisional instructions will be refined further once this configuration can be fully tested on the new clusters. RedHat Linux 6 for the two Deepthought clusters. 
| Uploader: | Gotaur |
| Date Added: | 5 October 2005 |
| File Size: | 7.91 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 27550 |
| Price: | Free* [*Free Regsitration Required] |
Verbs versions will not run on Cedar because of its different interconnect.
Please note that versions 2. Unfortunately, the Division of Information Technology does not have the expertise to assist in the assembling the inputs, etc. For best performance, NAMD jobs should use full nodes. In the results shown in first table below we used NAMD 2. This is due to the fact that nand compute nodes do not use AFS and so have copies of the AFS software tree, and so we only install packages as requested.
NAMD Hardware & Software Configurations | NVIDIA Data Center
RedHat Linux 6 for the two Deepthought clusters. This page was last edited on 21 Decemberat Skip to main content. Views Read View source View history. Pages with syntax highlighting errors Software. Now we perform benchmarking with GPUs. The numbers below were obtained for the standard NAMD apoa1 benchmark. Use the MPI version instead.
Usage guidelines and hints The following guidelines and hints only cover the mechanics of starting the namd2 program, especially on the High Performance Computing clusters. Simulation preparation and analysis is integrated into the VMD visualization package.
This section shows an example of how you should conduct benchmarking of NAMD. For packages which are libraries which other codes get built against, see the section on compiling codes for more help. If your run is too short, you might see fluctuations in your timing results.
Although we will continue to maintain these older versions of NAMD on the original Deepthought cluster for a while, they are getting long in the tooth and we encourage users to migrate one of the newer versions. Currently verbs versions do not work on cedar as they are incompatible with the communications fabric, so use MPI version instead.

NAMD is a parallel, object-oriented molecular dynamics code designed for high-performance simulation of large biomolecular systems. To do this, either: For a good benchmark, please vary the number of steps so that your system runs for a few minutes, and that timing information is collected in reasonable time intervals of at least a few seconds.
Since on graham GPU nodes your priority is charged the nmad for any job using up to 16 cores and 1 GPU, there is no benefit from running with 8 cores and 4 cores in this case. Using a verbs library is more efficient than using OpenMPI, hence only verbs versions are provided on systems where those are supported. The exact version is subject to change with little if any notice, and might be platform dependent.
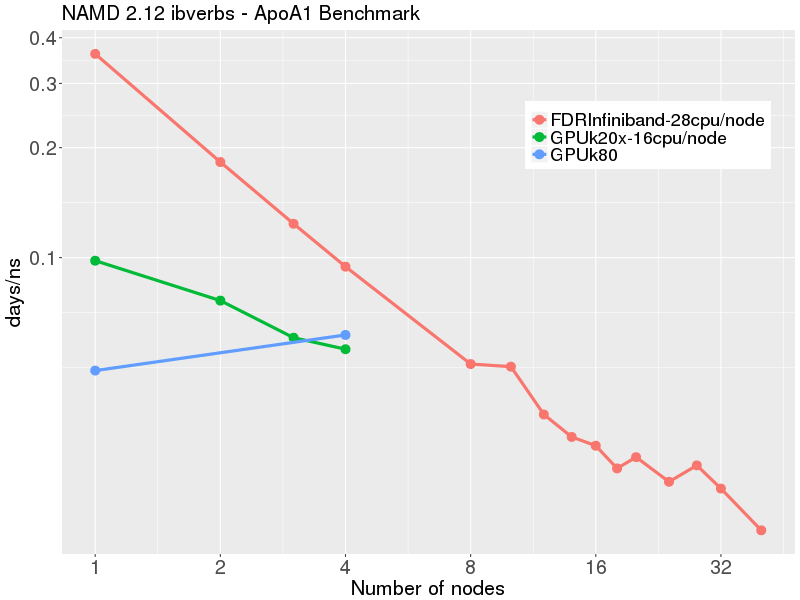
From our numbers we can see that using a full GPU node of graham 32 cores, 2 gpus jamd job runs faster that it would on 4 non-GPU nodes of graham. Navigation Wiki Main Page.
Compiling NAMD
Retrieved from " https: Finally, you have to ask whether to run with or without GPUs for this simulation. From this table it is clear that there is no point at all in using more than 1 node for this system, since performance actually becomes worse if we use 2 or more nodes.
You can also ask for details of how our NAMD modules were compiled. These results show that for this system it is acceptable to use up to cores. Therefore, if you plan to spend a significant amount of time simulating a particular system, it would be very useful to conduct the kind of benchmarking shown below. Packages labelled as "available" on an HPC cluster means that it can be used on the compute nodes of that cluster.
In general, you need to prepare your Unix environment to be able to use this software.

No comments:
Post a Comment